Unleash the Potential of CRISPR with Electroporation
By Michelle M. Ng, Ph. D.
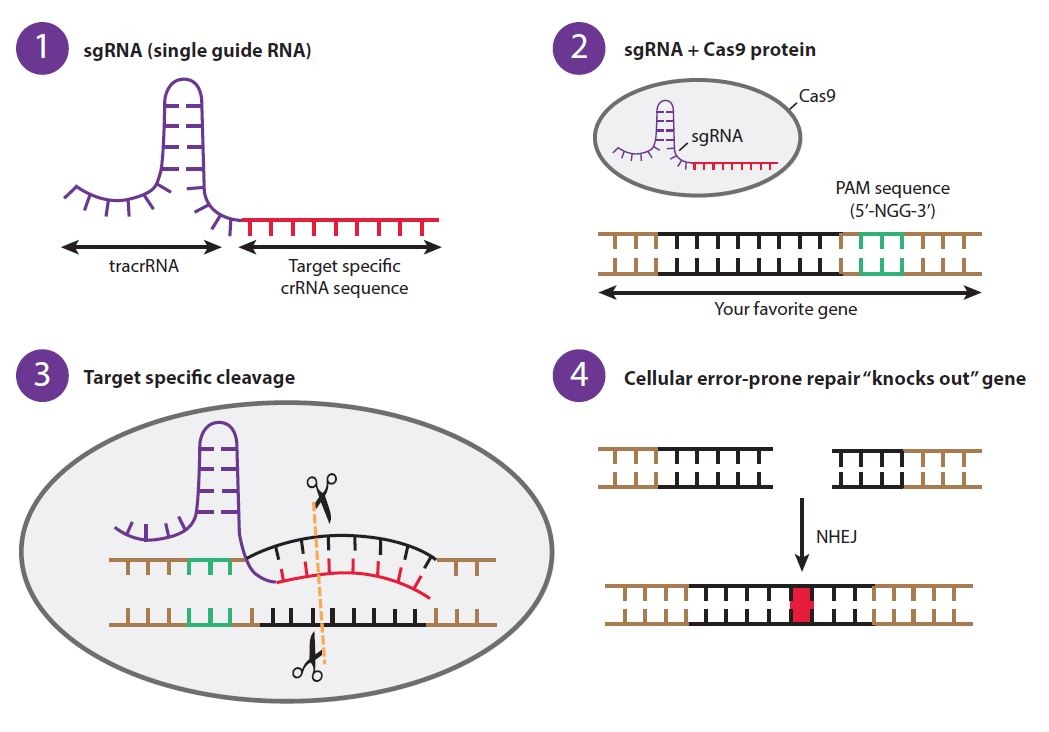
Part of this system has been modified to be used as a tool for editing genomes. A small guide RNA (sgRNA, 1) is employed which is complimentary to a PAM recognition site in the target area of the genome. Next the sgRNA and Cas9 protein form an ribonucleoprotein complex (2, RNP) which is capable of causing sequence-specific double stranded breaks at the PAM site (3).
These double stranded breaks can either be faithfully repaired or induce point mutations by DNA repair mechanisms of the cell (4, leading to frameshift mutation/knock-out) or be used to replace with DNA of interest at that site (Homology directed repair, HDR).
Here we explore the formats that CRISPR/Cas9 tools and the benefits of using electroporation to deliver CRISPR gene editing constructs.
What Forms of CRISPR Constructs? And Why?
- Plasmid based CAS9 and gRNA expressed from one construct. For this the expression promoters for the Cas9 protein and gRNA have to be compatible with your cell type. The plasmid lasts longest in the cell so the potential for off target effects are the greatest with this format. Because both the Cas9 protein and the gRNA have to be made and find each other in the cell to produce a functional RNP, this is also the least efficient format.
- mRNA , CAS9 and gRNA supplied in trans. This format is not cell type specific. mRNA format has an intermediate level of efficiency and has an intermediate half-life in the cell. Also the mRNA format has less chance of off target effects than plasmid format. Working with the mRNA can be daunting to some because RNA is sensitive to degradation.
- Protein CAS9 and gRNA precomplexed and delivered as an RNP. This is the most efficient method because the RNP is already ready to act once delivered to the cell and also is around for the shortest period of time (hours). Protein format also has the least risk of off-target effects
- Additional molecules for knock-ins: ssODN or dsDNA (PCR products or plasmid) for HDR template are added in addition to the CRISPR formats listed above. Short single stranded oligos (ssODNs) may be used for small changes such as a single amino acid substitution, however plasmids are most commonly used as HDR templates for larger knock-ins such as whole genes or florescent reporters.
Why Electroporation is an ideal delivery method for CRISPR
- Electroporation is an efficient method of delivering large or complex mixtures of molecules that were previously impossible to transfect by traditional chemical, lipid, and viral delivery methods. Multiplexed CRISPR gene editing and whole gene knock-ins are within reach with this technology.
- Electroporation is a physical method of transfection, and works well even with difficult to transfect samples, such as primary cells, suspension cells, embryonic cells, or even whole tissues in vivo. For 3D cultured cells or slices of tissue ex-vivo, the sample may be placed into a specialized electrode chamber. For in vivo transfection, the target tissue of the animal may be injected with the transfectant, then contacted with paddle shaped electrode probes, or reached by electrode needles inserted into the tissue.
- Electroporation pulse parameters are reproducible from one experiment to the next, and are not subject to lot to lot reagent variation or toxicity issues. The electrical pulses themselves are easy to monitor during the experiment using either an advanced electroporation instrument with pulse monitoring and logging features or attaching a high voltage probe and oscilloscope. Following electroporation, transient cell pores seal up leaving behind no residual chemicals that may affect cell viability.
Click here to learn more and download our latest CRISPR application note



 800-272-2775
800-272-2775
